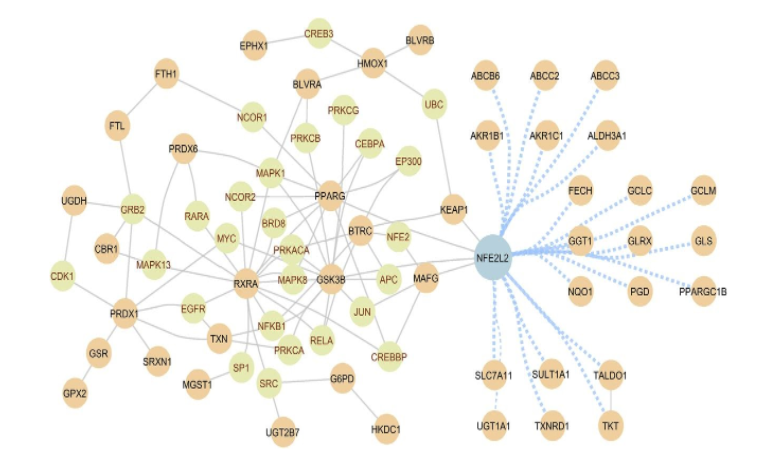
Locating the NRF2 regulatory pathway in the human interactome. NRF2 plays a key role in pathologic ROS formation as well as inflammatory and metabolic responses by coordinating the activity of various proteins. These interactions constitute a molecular interaction network, the NRF2 interactome. The known physical interactions between proteins involved in the NRF2 regulatory pathway (i.e., regulator proteins, brown circles) and the proteins through which they are connected (i.e., mediator proteins, green circles) are shown with a gray link. The regulator proteins that involve more than one mediator protein to connect to NRF2 are shown with a blue link. The regulator proteins are curated from literature, and their interactions are retrieved from various resources, including IntAct, MINT, BioGRID, HPRD, KEGG, and PhosphoSite (source, Cuadrado et al. Pharmacol Rev. 2018 Apr;70(2):348-383).
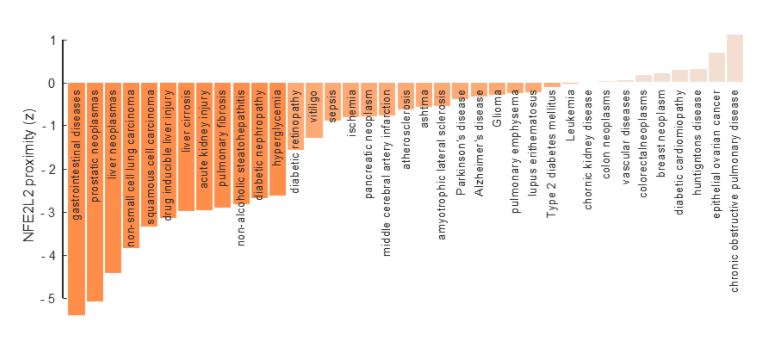
Systems medicine view of NRF2 from the perspective of the human interactome. Diseases are triggered by mutations that perturb proteins and their interactions, affecting a certain neighborhood in the NRF2 interactome. Interactome-based proximity measures the distance of genes to those disease neighborhoods. The bars show the proximity of NRF2 gene (NFE2L2) to known disease genes that participate in NRF2-related pathophenotypes. These bars highlight the role of NRF2 in digestive system diseases and cancers. For a given disease, the proximity first calculates the distance from NRF2 to the closest known disease gene and then compares that distance to a random expectation that is estimated using average distances between randomly selected proteins in the interactome. The z-score reported by interactome-based proximity corresponds to the significance of the observed distance between NRF2 and the disease genes. A negative value indicates that the observed distance is lower than what would be expected by chance. The bars are colored in varying tones of orange based on the z-score: significantly proximal (dark orange), proximal (orange), not proximal (light orange), respectively. The known disease genes are taken from DisGeNet, OMIM, and GWAS Catalog (source, Cuadrado et al. Pharmacol Rev. 2018 Apr;70(2):348-383).
COST (European Cooperation in Science and Technology) is a funding agency for research and innovation networks. Our Actions help connect research initiatives across Europe and enable scientists to grow their ideas by sharing them with their peers. This boosts their research, career and innovation.
Grant Holder: Universidad Autónoma de Madrid
Start of Action: 19 October, 2021
End of Action: 18 October, 2025
CSO approval date: 25 May 2021
Action email: info@benbedphar.org
© 2022 BenBedPhar | Design by Tuinbit Group